Contraction vs expansion of actomyosin networks in deep epithelia of drosophila
How to overcome a paradoxical situation in a complex biological context using dynamic Random Illumination Microscopy
Introduction: The challenge of dynamic tissue high resolution imaging
Tissue morphogenesis involves complex cellular rearrangements that depend on molecular dynamics at the nanometer scale, such as those of actomyosin networks. Imaging these processes in vivo and in real-time requires Super-Resolution microscopy (SR) that combines high spatial resolution, fast acquisition speed, and low phototoxicity, particularly for thick or scattering samples. Conventional SR methods are often limited by the complexity of their implementation, their phototoxicity, or their inability to image deep within samples. To overcome these constraints, Random Illumination Microscopy (RIM) has emerged as a robust and high-performing solution [2].
RIM: A robust super-resolution modality for live imaging
RIM is a super-resolution technique based on illuminating the sample with a series of random light patterns (speckles), generated by a binary phase modulator (Spatial Light Modulator – SLM). These random patterns encode high spatial frequency information into the observed images. The software then performs a statistical reconstruction from multiple low-resolution images to generate a final image with doubled resolution (approximately 100 nm laterally) and robust against artifacts.
Key Advantages of RIM:
-
-
- High resolution for dynamic imaging: Offers resolution comparable to Structured Illumination Microscopy (SIM).
- High penetration depth and insensitivity to aberrations and scattering: Unlike SIM, RIM is minimally affected by optical aberrations, making it particularly effective for deep imaging in thick, live tissues, such as embryos or organs.
- Low phototoxicity and high speed: Allows for rapid and repeated acquisition with a reduced light dose, which is essential for tracking cellular and molecular dynamics over long periods. In the context of the Drosophila study [1], 3D + time RIM was performed with a temporal resolution of 20ms to track actomyosin networks.
-
Contraction vs expansion: How to overcome a paradoxical situation in biological context
Organ and tissue shape is largely driven by cytoskeletal forces, especially actomyosin networks. Traditionally, these networks are contractile: they shrink cell surfaces and help deform tissues during processes such as invagination, constriction, or closure. However, in this use case study by Wang et al., the authors revealed a paradoxical situation:
During late Drosophila oogenesis, follicle epithelial cells must rapidly expand their basal surface to match the fast growth and elongation of the oocyte during nurse-cell dumping. Yet these same cells contain dense basal stress fibers enriched in Myosin-II, normally expected to contract, not expand, the cell surface.
In this study, the authors used RIM modality (LiveDRIM system) to reveal actin/myosin nanostructures and their dynamics, which informed on the distinct nano-structure of expanding stress fibers and F-actin/myosin signal separation, from traditional contractile stress fibers.
Figure 1. LiveDRIM system setup
How can cells and tissues expand when their cytoskeleton is built from contractile actomyosin fibers?
The study shows that follicle cells turn their basal actomyosin network into an unusual, non-contractile and expanding structure, with pulsatile dynamics, non-linear architecture, and spatial separation of actin and Myosin-II. These subtle and fast events could only be revealed using RIM (Random Illumination Microscopy), thanks to its combination of super-resolution, deep imaging, low phototoxicity, and fast acquisition. Indeed, the key biological discoveries rely on the ability to observe:
- sub-100–150 nm actin and Myosin-II structures,
- fast cytoskeletal pulses and focal adhesions dynamic,
- deep tissue basolateral surfaces,
- long term time-lapse sequences (1–3 hours).
RIM modality uniquely provides all these capabilities simultaneously.
Superior resolution to reveal the “non-linear” actomyosin architecture
RIM achieves 100 nm lateral resolution without the limitations of classic structured illumination.
This allowed the authors to visualize fragmented actomyosin fibers, branched networks, intermittent Myosin-II enrichments and decoupled actin/Myosin-II spatio-temporal dynamics. These nanoscale features are essential to demonstrate that the network is non-contractile. They are not detectable with standard confocal or spinning-disk imaging.
Deep-tissue imaging: Accessing the basal side of a curved epithelium
The Drosophila follicular epithelium forms a thick, curved tissue (Fig. 2 a., b.), and the structures of interest lie at the basal surface, behind multiple scattering layers. Conventional super-resolution approaches (SIM, Airyscan) degrade rapidly in such conditions.
RIM modality maintains high resolution and signal-to-noise even at depth:
- illumination speckles are robust to aberrations,
- statistical reconstruction compensates for optical distortions,
- no precise illumination pattern needs to be preserved
This made it possible to map actomyosin organization across the entire epithelium during the ruffling, increasing, and spreading phases.
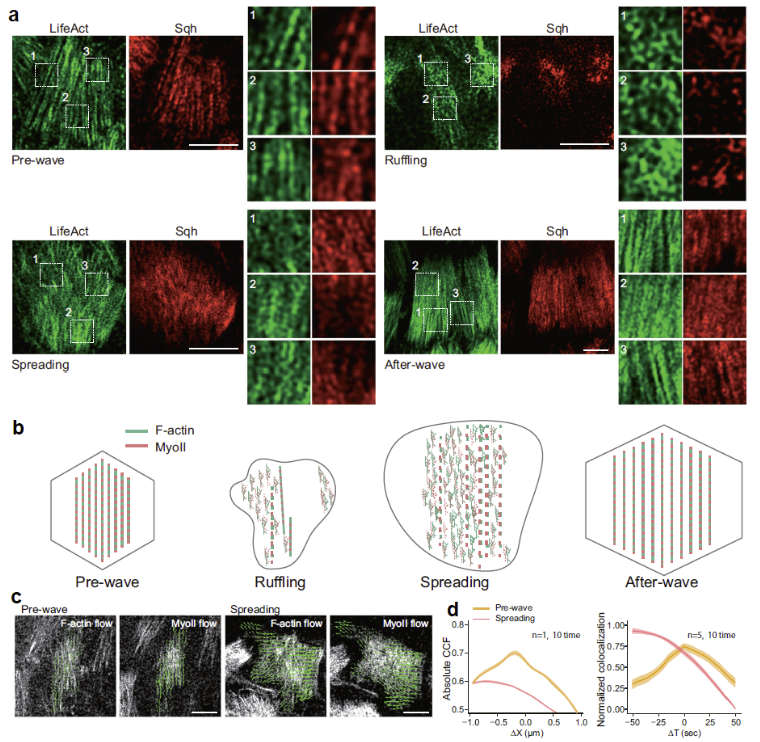
Figure 3. a. Nanoscale structures of basal actomyosin networks in follicle cells during the indicated phases, including raw images and ROI enlarged images of F-actin and Myosin-II signals. b. Representative cartoons summarizing linear and non-linear patterns of basal actomyosin networks before, during and after epithelial expansion behaviour. Green and red coolors mark F-actin and Myosin-II signals, respectively. c. Nanoscale fiber dynamics of basal F-actin and Myosin-II signals in follicle cells during the pre-wave and spreading phases. The arrow direction indicates the signal flow orientation, and the length indicates the relative velocity. d. Spatial (left) and temporal (right) correlation analyses of basal F-actin and Myosin-II signals in follicle cells during the pre-wave and spreading phases. Data are presented as mean values +/- SD. Scale bars are 5µm.
Low phototoxicity: Critical for 3-hour live imaging
The biological system is extremely sensitive. Excessive light alters actomyosin dynamics, disrupts oocyte growth, blocks nurse cell dumping and affects cell migration and spreading.
RIM uses low-intensity illumination (random speckle patterns) avoiding hot spots, and reconstruction-based contrast enhancement instead of higher light dose. Long-term live imaging (up to 3 hours) was achieved without inducing artefacts. The natural pulsatile behavior and the mechanical coupling with the growing oocyte could be captured faithfully [1].
In the study by Wang et al. (Fig. 3), 3D + Time RIM was used to reveal the nanoscopic structure and dynamics of basal actomyosin networks in Drosophila follicle cells. This analysis made it possible to characterize the transition from connected (contractile) structures to disconnected and non-linear (non-contractile/expansive) structures during the expansion phase.
Fast acquisition: Capturing rapid pulses and adhesion dynamics
RIM enables high-speed image acquisition (tens to hundreds of ms) while retaining super-resolution after reconstruction [3]. This is essential since:
- actomyosin pulses occur every 30–120 s,
- myosin-II and actin flow at ~0.2–0.3 µm/min,
- focal adhesions assemble/disassemble rapidly (time scale of seconds),
- spreading waves propagate along the tissue.
This study is an excellent demonstration of how Random Illumination Microscopy (RIM) and our technology LiveDRIM enables biological insights that are otherwise inaccessible in vivo. RIM modality allowed the researchers to uncover an entirely new mode of actomyosin behavior: an expanding, non-contractile network that enables epithelial cells to flatten and elongate during morphogenesis.
RIM therefore appears as a uniquely powerful tool for studying rapid cytoskeletal dynamics in thick, living tissues.
INSCOPER: Optimal control for RIM experiments
The INSCOPER software is the backbone that guarantees the optimal performance of the RIM system. Developed for universal and latency-free control of fluorescence microscopes and their third-party peripherals, INSCOPER is essential for the precise synchronization required by RIM.
- Precision Synchronization: In this study, RIM acquisition requires the capture of multiple images under distinct speckle illuminations. INSCOPER ensures the necessary synchronization between the sCMOS camera, the laser source, and the SLM, guaranteeing the integrity of the raw super-resolution data acquisition.
- 3D + Time (4D) Acquisition: 3D + Time RIM was used to track the subtle dynamics of actin filaments (LifeAct-GFP) and Myosin-II (Sqh-RFP). INSCOPER’s latency-free control maximizes the acquisition rate, ensuring that the rapid dynamics of actomyosin flow (average speed up to 0.3 μm/min during the spreading phase) are captured with optimal temporal fidelity.
- System Flexibility: The LiveDRIM system (Dynamic Random Illumination Microscopy) developed by Rimeo and distributed by INSCOPER and Gataca-Systems can be used regardless of the hardware, as each setup can potentially be customized. INSCOPER is compatible with all third-party devices, allowing control of complex configurations (such as the use of multiple cameras) and easy integration of fast laser modulation (e.g., Oxxius) and a binary phase modulator (SLM) used in the study.
This application note was written with the agreement of the corresponding authors of the cited article Li, S. et al. Basal actomyosin pulses expand epithelium coordinating cell flattening and tissue elongation. Nat. Commun. 15, 3000 (2024).
Bibliography
- Li, S. et al. Basal actomyosin pulses expand epithelium coordinating cell flattening and tissue elongation. Nat. Commun. 15, 3000 (2024).
- Mangeat, T. et al. Super-resolved live-cell imaging using random illumination microscopy. Cell Rep. Methods 1, 100009 (2021).
- Labouesse, S. et al. Pseudo Random Illumination for Live Super-Resolution Microscopy. (2025).


0 Comments