Cutting-edge technology offering near-instant deconvolution
by seamlessly integrating intelligent software programming
with the computational power of a GPU
Comparison of the raw image (left) with the Microvolution processed image (right) in the Inscoper I.S. environment.
Technique: Widefield
Slide: mouse kidney section with Alexa Fluor 488 WGA, Alexa Fluor 568 phalloidin, DAPI
Microvolution® is a software dedicated to 2D and 3D image deconvolution originally developed by Stanford scientists.
Microvolution benefits
Improved visibility in low light conditions, increasing success rates in live cell and time-lapse experiments.
Improved Measurement Accuracy: cleaner measurements after deconvolution, including improved colocalization, FRET data, neurite lengths, and fluorescence intensities.
Dynamic Experiment Adjustments: Ability to make real-time adjustments to microscopy experiments, allowing more data to be collected from the same sample.
Cross-Microscope Compatibility: Microvolution software deconvolves images from widefield, confocal, two-photon, light sheet, and HCA microscopes.
Microvolution technology uses blind deconvolution to improve noisy data, especially in deep tissue imaging.
Multi-GPU options allow fast processing of even very large samples.
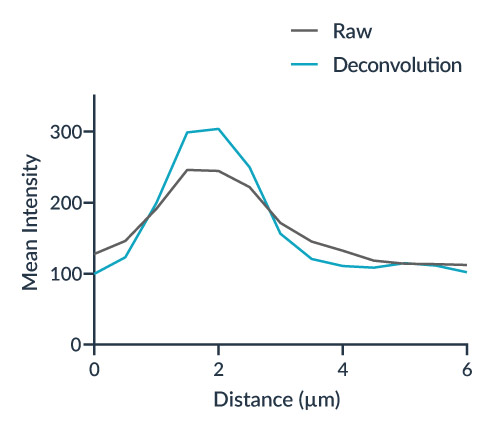
Accuracy: Microvolution preserves thin filaments using a precise Richardson-Lucy algorithm, ensuring accuracy up to 200 times faster than other software vendors.
Clarity & Increased Resolution: Deconvolution under optimal conditions breaks the diffraction barrier, improving visual resolution. Microvolution’s technology is demonstrated with a 2X improvement in clarity.
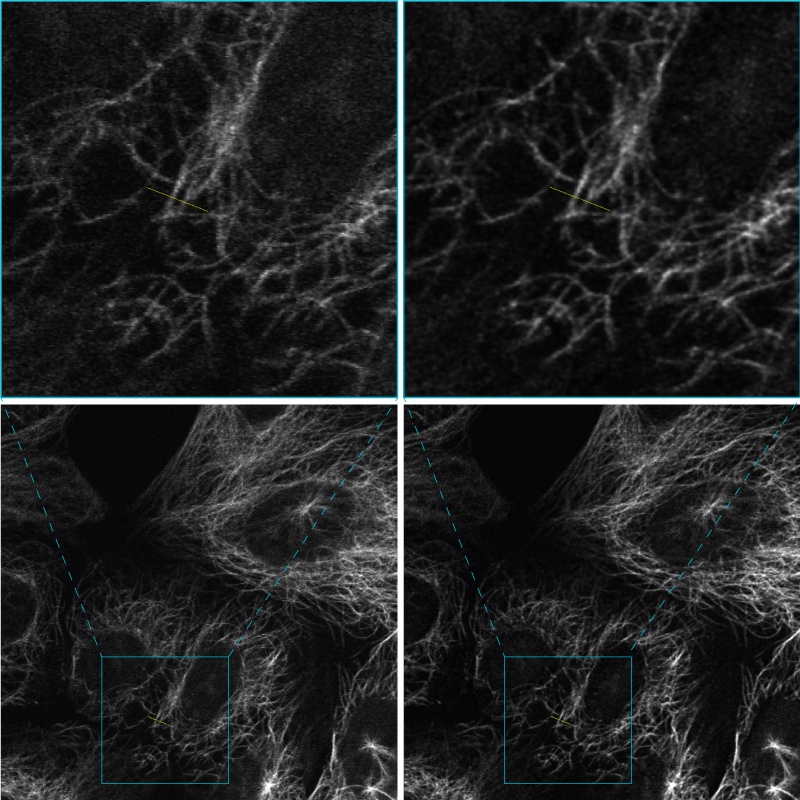
Before and after deconvolution example
Raw (left) and deconvolved (right) image: cells labeled with microtubules, Alexa fluor 647. Image courtesy of Dr. Stéphanie Dutertre, MRic (Microscopy Rennes Imaging Center) Université de Rennes 1 SFR Biosit (UMS 3480 – US 018)
demonstration of the Microvolution software
integrated in the Inscoper I.S. image acquisition workflow
image gallery
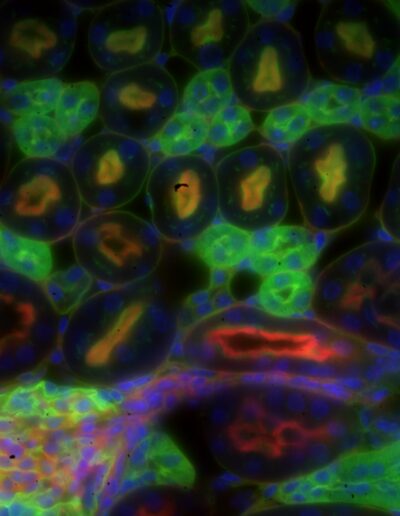
Mouse kidney section with Alexa Fluor 488 WGA, Alexa Fluor 568 phalloidin, DAPI – Widefield – Raw image
Image by Dr. Claire Déméautis – INSCOPER
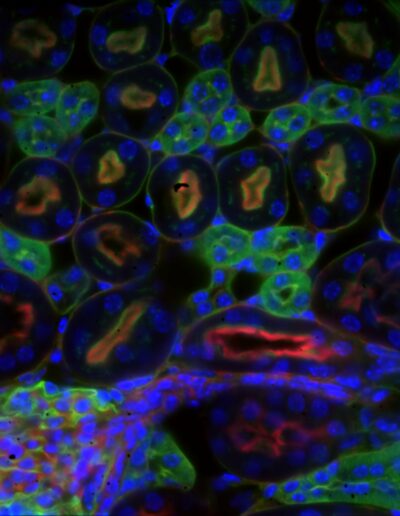
Mouse kidney section with Alexa Fluor 488 WGA, Alexa Fluor 568 phalloidin, DAPI – Widefield – Deconvolved image
Image by Dr. Claire Déméautis – INSCOPER
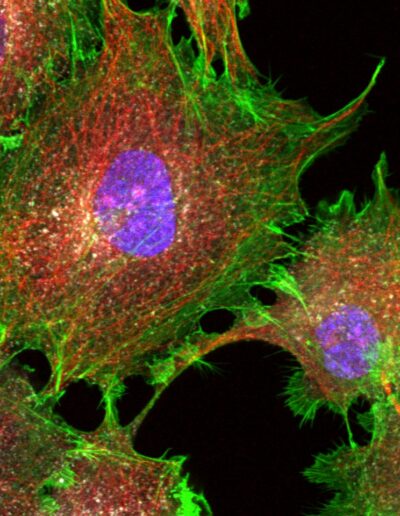
COS cells labeled for DNA (DAPI, 405), actin (phalloidin, 488), microtubules (immunolabeling, 561) and clathrin (immunolabeling, 647) – Confocal – Raw image
Image courtesy of Dr. Christophe Leterrier, NeuroCyto team, INP CNRS UMR 7051 – Aix Marseille Université
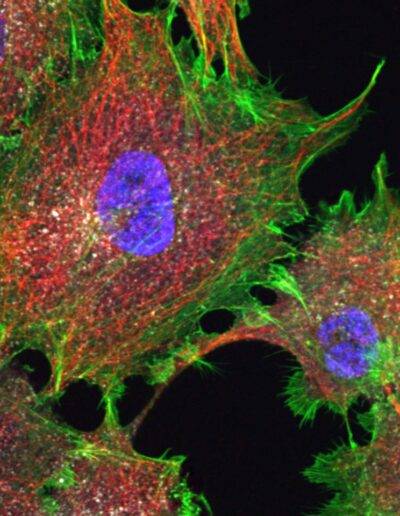
COS cells labeled for DNA (DAPI, 405), actin (phalloidin, 488), microtubules (immunolabeling, 561) and clathrin (immunolabeling, 647) – Confocal – Deconvolved image
Image courtesy of Dr. Christophe Leterrier, NeuroCyto team, INP CNRS UMR 7051 – Aix Marseille Université
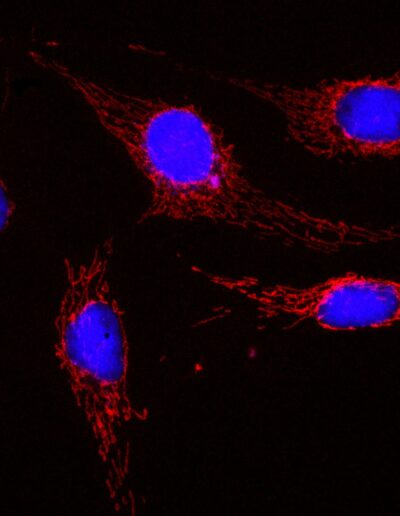
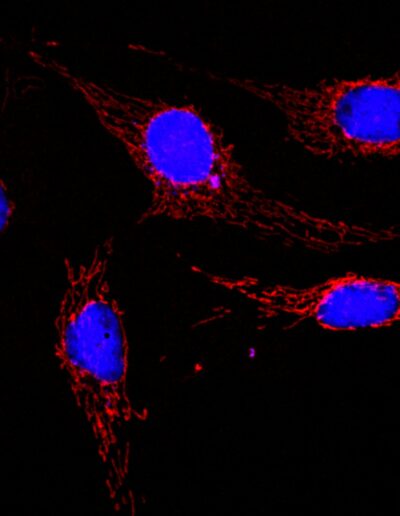
BPAE cells with MitoTracker red CMXRos deconvolved image (crop)
Image by Dr. Claire Déméautis – INSCOPER