Maintenance of the genome integrity is directly dependent on the spatiotemporal recruitment and regulation of the repair proteins at DNA damage sites. The alteration of this complex biological pathway could induce mutations or premature cell death. The real-time monitoring of this phenomenon can be done using a technique called laser microirradiation illumination with highly energetic photons focused towards the desired area to induce damages. Such an approach has become a powerful tool to explore the DNA repairment following laser-induced damage. This application note introduces the use of Inscoper scanFRAP solution to explore the kinetic of protein recruitment following laser-induced DNA damage.
Biological context
The integrity of genomic DNA is continually challenged by the exposition of genotoxic factors such as radiations, chemicals, reactive products of cellular metabolism or environment mutagens. One of them is ultraviolet light (UV) that can induce 100,000 DNA lesions per hour per exposed cell (Jackson & Bartek, 2009). All of them could lead to DNA single or double strands breaks. If they remain unrepaired, these pathological phenomena could induce mutations and subsequently cancer and/or cell death. To prevent these alterations, cells have developed an efficient tool to repair DNA and restore its integrity as fast as possible.
Laser microirradiation can induce artificial DNA lesions to evaluate the recruitment of a large panel of repair enzymes (Figure 1). Used for the first time in 1980 (Cremer et al, 1980), it has become nowadays a key technique to study the involvement of DNA repair (Suzuki et al, 2011; Zentout et al, 2021; Kong et al, 2021).
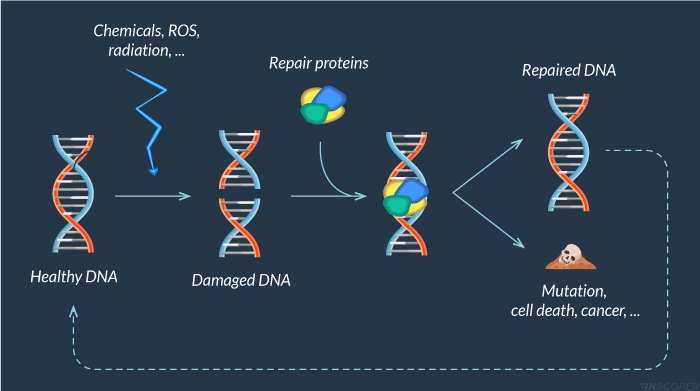
Figure 1: DNA damage response
Cells are continuously faced with exogenous (chemicals, UV light, ionizing radiations) and endogenous (ROS, …) agents that can lead to DNA damage. When DNA is damaged, a large panel of proteins is recruited to repair this alteration. Unrepaired lesions could alter DNA integrity and lead to premature cell death, cancer or mutations.
Microirradiation with the Inscoper scanFRAP
Inscoper scanFRAP is a complete microscopy solution for photomanipulation and optogenetics. The product consists of a software and hardware package compatible with advanced video microscopes used in life science. Incorporating a specially-designed electronic unit to control the microscope stand and third-party devices, the Inscoper scanFRAP provides a new user experience for photomanipulation applications with improved technical performance, full system integration and ease of use.
Users benefit from a state-of-the-art solution to add photomanipulation and optogenetics experiments within their conventional acquisition sequences, allowing a full and smooth image workflow compatible with the other imaging techniques also implemented on their system. The core of Inscoper technology eliminates any software latency when controlling the overall microscopy system. It increases the temporal resolution, compared to conventional approaches, which is a major advantage for applications in live cell imaging (Figure 2).
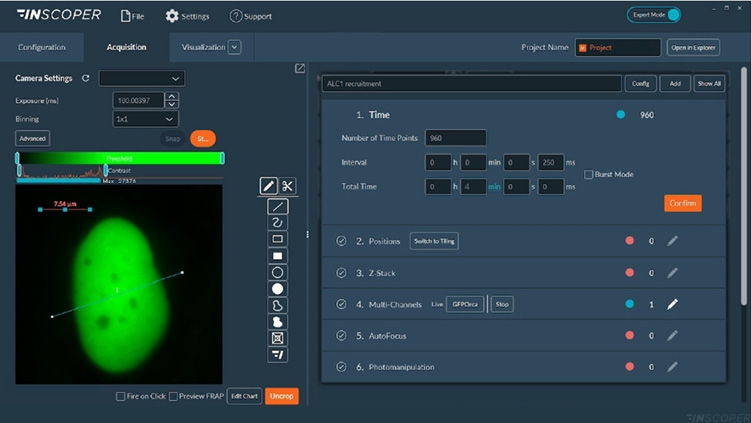
Figure 2: Interface of the Inscoper software
Overview of the Inscoper software used to manage the acquisition sequence with multidimensional parameters including timelapse, multiposition or/and tiling, Z-stack, multi-channels and photomanipulation for microirradiation, and FRAP applications. All of these dimensions are fully customizable to be more suitable for user’s experiments.
Based on galvanometric mirror technology, the Inscoper scanFRAP is a microscopy solution for photomanipulation applications including FRAP, photoactivation or microirradiation. Here, this application note only focuses on microirradiation application. Researchers have full control of all laser settings. They can completely personalize and optimize the microirradiation areas, modulating in live the region of interest (ROI) in the software user interface. Faster acquisition speed allows to monitor biological phenomena after microirradiation with a very high temporal resolution, such as the recruitment of repair proteins following laser-induced damage.
Microirradiation experiments
Objectives
Here, we want to characterize the recruitment of the ALC1 (chromodomain-helicase-DNA-binding protein 1-like) protein following laser-induced DNA damage. This protein is involved in DNA unpackaging to allow the recruitment of other repair enzymes (Sellou et al, 2016).
Material
A Nikon Ti2 Eclipse microscope (Nikon, Tokyo, Japan) with a Plan Apo λ 60x 1.4 NA oil immersion objective (MRD01605; Nikon) was used. For these experiments, the camera was a digital CMOS ORCA-Fusion BT (C15440-20UP; Hamamatsu Photonics, Hamamatsu, Japan) and the light engine was from Lumencor (SpectraX; Lumencor, Beaverton, USA). Microirradiation was performed using Inscoper scanFRAP (INSCOPER, Cesson-Sévigné, France) with a 405nm laser source (L6Cc; Oxxius, Lannion, France).
Method
U2OS cells were transfected to overexpress ALC1 fused with GFP (Green Fluorescent Protein), resulting in an ALC1-GFP protein (GenBank AF537213.1,A). Nuclei were labeled using Hoechst 33342 (H3570; ThermoFisher Scientific, Waltham, USA) before imaging. For this experiment, the microirradiation was induced using the 405nm laser (100% intensity for 1 second) to induce DNA damages and the recruitment of ALC1-GFP protein at the injured area. This measurement allows to characterize the ALC1 protein dynamic, following laser-induced DNA damage. ALC1-GFP recruitment was monitored using a 488 nm excitation (exposure time: 100 ms and frame rate: 4fps).
Results
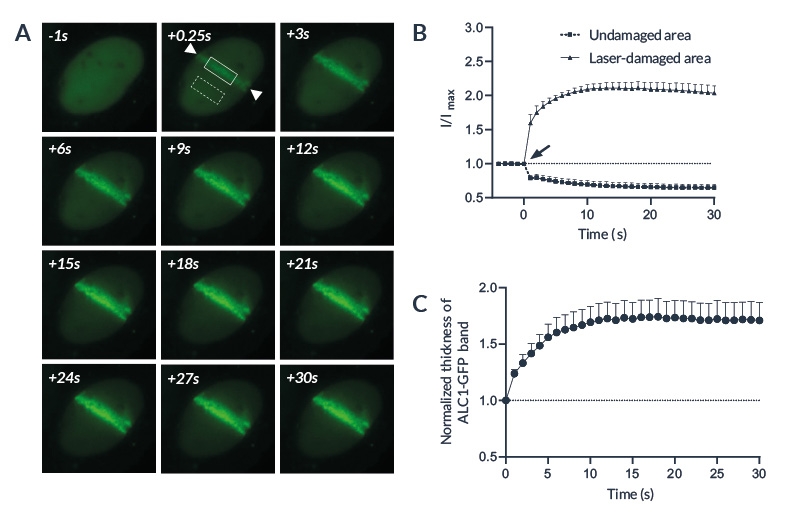
The laser microirradiation represents a powerful tool to monitor DNA repair with high temporal and spatial resolution. DNA lesions were induced following ROI (region of interest) previously defined using the Inscoper software. During the acquisitions, a progressive recruitment of ALC1-GFP protein could be observed over the entire length of the ROI (Figure 3A). Mean intensity of the fluorescent signal has been then measured on the damaged area. The signal from an unaltered ROI was also measured as control. A rapid increase of the signal was observed in the irradiated area, to remain stable in less than 10 seconds (Figure 3B). On the contrary, intensity from the control ROI appeared slightly decreased following the photomanipulation. ALC1-GFP accumulation could also be characterized by the width of the band (Figure 3C).
Figure 3: ALC1 recruitment following laser-induced DNA damage
(A) Representative image showing ALC1-GFP accumulation at sites of laser-induced DNA damage in U2OS cells. Area between the arrows has been damaged by a 405nm laser. The solid and the dashed boxes represent respectively the laser-damaged and the control area. (B) Quantified accumulation of ALC1-GFP following microirradiation (arrow) on laser-damaged (circles) and undamaged (squares) areas. (C) Normalized thickness of the ALC1-GFP band in the irradiated region following laser-induced DNA damage
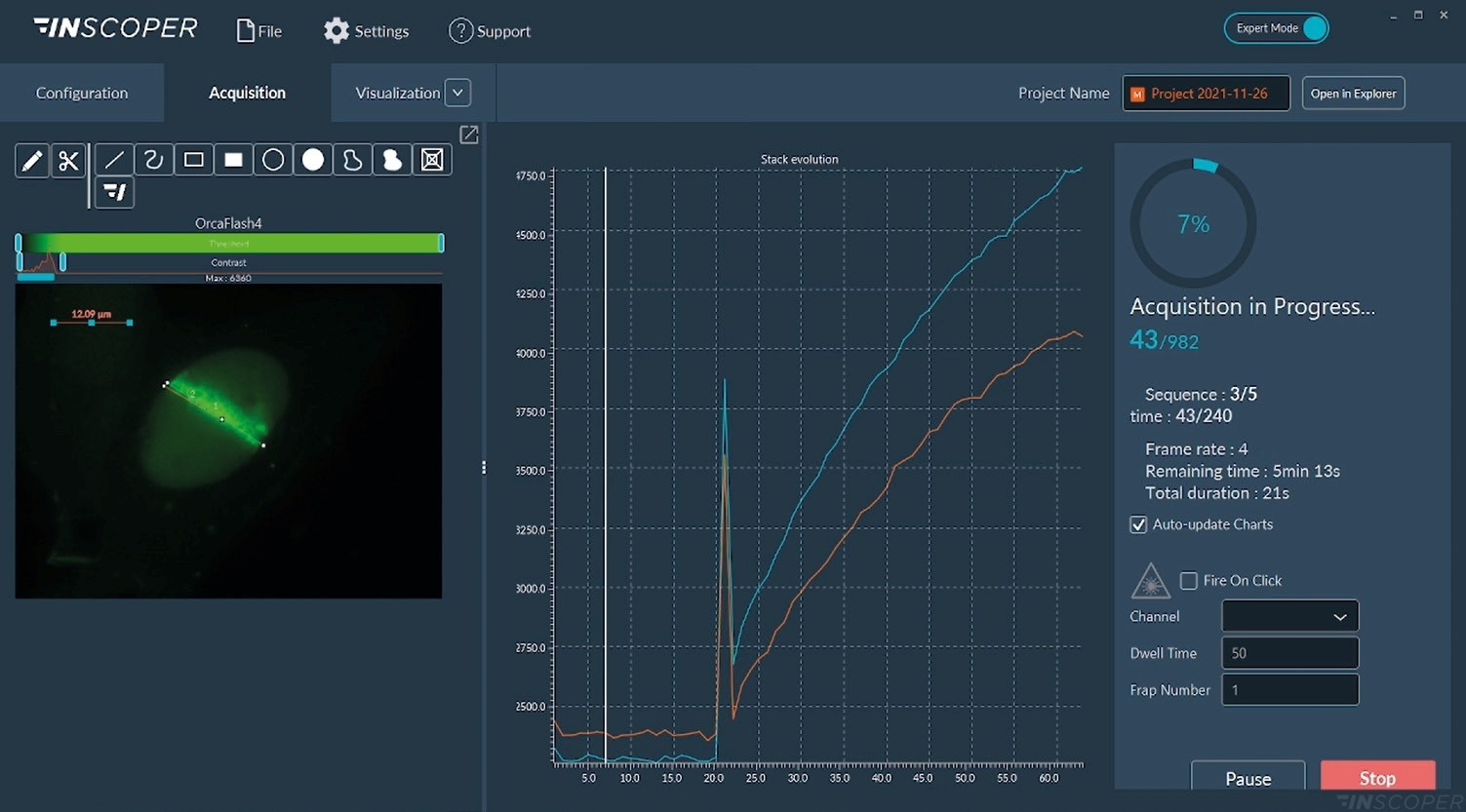
Figure 4: Interface view of the Inscoper software during FRAP acquisition
Acquisitions are real-time monitored with images or graphics. The “Fire-on-Click” (bottom right part of the interface) can be activated at any moment to add a new ROI to photobleach while the acquisition is running.
Summary
The laser microirradiation on living cells helps to study the cellular response to DNA damages. The use of the Inscoper scanFRAP solution allows to monitor in real-time the recruitment of a wide spectrum of DNA repair proteins with high spatiotemporal resolution. It could also be useful to evaluate the impact of drugs on the maintenance of the DNA integrity. Laser microirradiation could be combined with other microscopy approaches to better characterize biological pathways involved during DNA repair. For instance, it could be coupled with FLIM (Fluorescence Lifetime Imaging Microscopy) to evaluate the cellular metabolic changes (Murata et al., 2019) or FRET (Fluorescence Resonance Energy Transfer) to characterize the kinetics of protein interactions (Lou et al., 2019).
Bibliography
- Cremer C, Cremer T, Fukuda M & Nakanishi K (1980) Detection of laser-UV microirradiation-induced DNA photolesions by immunofluorescent staining. Hum Genet 54: 107–110
- Jackson SP & Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461: 1071–1078
- Kong X, Wakida NM & Yokomori K (2021) Application of Laser Microirradiation in the Investigations of Cellular Responses to DNA Damage. Front Phys 8: 1–8
- Lou J, Scipioni L, Wright BK, Bartolec TK, Zhang J, Masamsetti VP, Gaus K, Gratton E, Cesare AJ & Hinde E (2019) Phasor histone FLIM-FRET microscopy quantifies spatiotemporal rearrangement of chromatin architecture during the DNA damage response. Proc Natl Acad Sci 116: 7323–7332
- Murata MM, Kong X, Moncada E, Chen Y, Imamura H, Wang P, Berns MW, Yokomori K & Digman MA (2019) NAD+ consumption by PARP1 in response to DNA damage triggers metabolic shift critical for damaged cell survival. Mol Biol Cell 30: 2584–2597
- Sellou H, Lebeaupin T, Chapuis C, Smith R, Hegele A, Singh HR, Kozlowski M, Bultmann S, Ladurner AG, Timinszky G, et al (2016) The poly(ADP-ribose)-dependent chromatin remodeler Alc1 induces local chromatin relaxation upon DNA damage. Mol Biol Cell 27: 3791–3799
- Suzuki K, Yamauchi M, Oka Y, Suzuki M & Yamashita S (2011) Creating localized DNA double-strand breaks with microirradiation. Nat Protoc 6: 134–139
- Zentout S, Smith R, Jacquier M & Huet S (2021) New Methodologies to Study DNA Repair Processes in Space and Time Within Living Cells. Front Cell Dev Biol 9: 1–15

0 Comments